Gene Trees now have intron ticks.
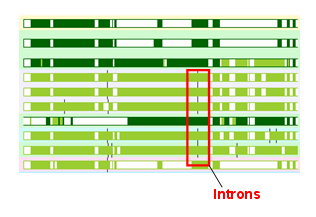
The gene tree images now have little intron "ticks" on them showing how the intron position is placed relative to the protein sequence. An example is shown above. Each tick is a little black line on each side of the green protein bars, on the right. As intron positions have been remarkably stable on the "chordate" side of the metazoan tree (ie, the deutrosomes), one should expect that the introns line up - if they do, it is good evidence that the alignment is right.
There are some interesting things. Ensembl models small frameshifts to create open reading frames around erroneous data as tiny introns. In this code you cannot distinguish these two classes of introns, but as these errors normally come in patches, a run of intron ticks unique to a genome is probably a set of errors (an example is in Gorilla). I've enjoyed browsing around some of my favourite genes to check out that the introns make sense.
There is some more to go here. The fact that the intron ticks disappear on collapsed nodes is a bit frustrating - it would be nice to see "consensus" intron positions (though this is a bit complex to execute underneath).
2 comments:
If Ensembl "models small frameshifts to create open reading frames around erroneous data as tiny introns", is there a way to know whether these occur due to errors in the data or if these frameshifts really indicate a pseudogene?
This is a good question. The small introns can be indicators of an erroneous cDNA that does not align 100% to the genome, or a problem with the genomic assembly itself. Or, the transcript may truely not be transcribed and expressed.
To investigate this, one can look at the supporting evidence on which a transcript was based. Click through the diagrams to see the cDNA and/or protein used to determine the transcript. This can indicate if the small introns allowed all records to align (in which case it is more likely a problem with the genomic sequence) or if the evidence itself has a frameshift mutation. Protein evidence and ESTs in the region would back up a "real" protein-coding transcript, as long as they do not also align elsewhere. These tracks can be turned on in the location tab, using the configure this page button at the left.
Finally, to ascertain the quality of a genomic region, for human have a look at the 1000 genomes browser. Comparing the region across multiple individuals can indicate trouble areas of the reference genome. Be aware the coordinates are for the older NCBI36 assembly. To directly compare with the newer GRCh37 assembly, you can use the Ensembl assembly converter.
Post a Comment